Amie Adkin gives an update on a recent workshop for the COMPARE project that was by all accounts beyond compare.
What is COMPARE?

The world is increasingly facing outbreaks of infectious diseases in wildlife and livestock. Some of these diseases have the potential to cause illness in people, for example if they slip through our biosecurity controls within the food-chain, into our kitchens, and onto our plates.
Wildlife, such as migratory birds, pay no attention to national borders and can facilitate the spread of infectious diseases between the livestock residing in different countries. This means that a common approach to investigating and dealing with disease outbreaks across nations will speed up the identification of what is causing the outbreak, and help to minimise the risk posed to livestock and the general public.
 COMPARE is a multidisciplinary research network with major contributions from a whole host of European nations. It is an ingenious acronym of COllaborative Management Platform for detection and Analyses of Re-emerging and foodborne outbreaks in Europe.
COMPARE is a multidisciplinary research network with major contributions from a whole host of European nations. It is an ingenious acronym of COllaborative Management Platform for detection and Analyses of Re-emerging and foodborne outbreaks in Europe.
COMPARE was set up with the common vision to create a standard framework for analysing data from genetic sequencing of pathogens. The aim is to form a platform for globally linked data to aid the rapid identification, containment and mitigation of emerging infectious diseases and foodborne outbreaks (http://www.compare-europe.eu/).
The 5 year project has been awarded €20 million funding under the European Commission’s Horizon 2020 programme. APHA are contributing in all work areas of: human health, livestock health, wildlife and food safety, which encourages joined-up work-flows and networks fostering “one health” thinking across the consortium.
The workshop: an outbreak of ideas
Now at the mid-point of the project, APHA hosted a workshop at Weybridge with representatives from many key stakeholders including the Department for Environment Food & Rural Affairs (Defra), the Veterinary Medicines Directive (VMD), Public Health England (PHE) and the Food Standards Agency (FSA).
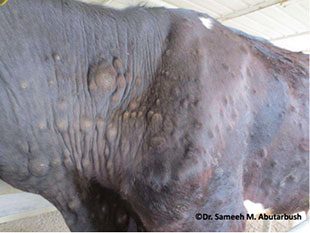
A presentation was given on recent advances in developing a generic risk assessment to assist in targeting surveillance, using Lumpy skin disease as a case study which is an exotic livestock disease. It highlighted those areas which are at most risk in the UK and across Europe from importing infected live animals.
Other results hot off the press were presented on work with the avian influenza virus ‘H5N8’. Analysis of the genetic sequence of this virus helps researchers understand how changes in the virus affects the bird species that can be infected such as chickens and turkeys, which links to the role of wildlife in recent disease outbreaks. Such work aids researchers understand why certain host populations have been more likely to become infected than others.
Research has also explored different approaches to early detection of outbreaks. For instance, the analysis of genetic sequences using ‘cluster detection algorithms’ to trace the source of an outbreak, has recently been tested in an outbreak of Salmonella Typhimurium.
A key principle of the COMPARE project is that sharing genetic sequence data will enable the rapid detection of disease outbreaks. However, this will only work effectively if all data is shared as soon as it is generated. Is this achievable within the current regulatory framework? Could a platform encourage the faster sharing of data whilst also ensuring countries’ individual confidentially requirements are met? Discussion of this topic acknowledged that confidentiality agreements would need to be carefully considered with potentially different levels of data access.
The potential for the COMPARE platform to combat re-emerging and foodborne outbreaks in Europe is going from strength-to-strength. We are already looking forward to presenting and highlighting future findings at our next meeting, such is the fast pace of ongoing work.
Visit the COMPARE website to find out more.

1 comment
Comment by Daniel John posted on
Nice work guys