
March 2nd marked the 9th International Open Data Day, an annual event promoting awareness and use of open data. Colin Birch, lead statistician at APHA, shares with us his first time experience of publishing a disease simulation model by open access and public release.
What is open data?
Open data can be defined as “data that anyone can access, use or share” (Open Data Institute definition). Traditionally, access and re-use of data has been controlled by public and private organisations through access restrictions, copyrights and patents, and would often incur charges. While the debate on open data is still ongoing, there has been an increased acceptance that information created by a government institution using public money should be made universally available. In 2010, data.gov.uk was launched and houses over 30,000 public sector data sets, from cross-border pig movements to the estimated cost of organised crime!
These 'open government initiatives' have arisen out of a growing citizen expectation of the role that governments play in facilitating public transparency. In parallel, we have observed this past decade an increasing demand by the general public to be allowed to engage with decision making through the unrestricted use, modification and re-distribution of the necessary data and tools. Publicly releasing government models is another logical step in facilitating active public participation in policy analytics.
Modellers and others trying to publish work on complex applications often experience a Catch 22. To publish a model requires evidence that the model is fit for purpose, but to publish evidence that the model is fit for purpose requires a reference explaining the model. A model description is dense, challenging reading that will appeal to only a small audience. Model development involves decisions about the priorities for the model’s simplification, so a thorough description may provide many opportunities for critics to disagree with authors. Hence it is notoriously difficult to publish scientific papers on simulation models.
Open access and public release offer a solution to this challenge. Journals are increasingly keen for authors to provide as much information and data as possible to allow readers to check and reproduce reported results. Online publishing removes all limits on the size of the supplementary material that can be associated with a paper. In modelling, the ultimate response is to provide the model, not just a description, with a paper presenting its outputs. Debates with referees about how a model will behave become obsolete when the model itself is generally available for testing. I decided to attempt this approach when I submitted a paper to BioMed Central (BMC) Veterinary Research on outputs from a previously unpublished model on bovine tuberculosis.
The Bovine Tuberculosis Model for England and Wales
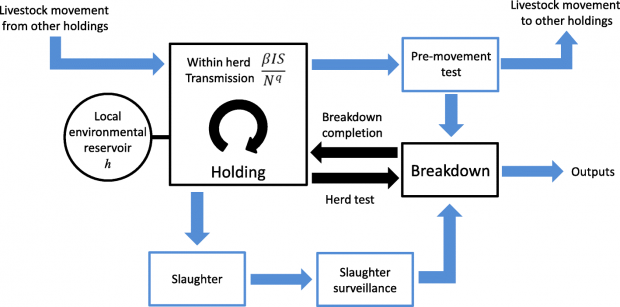
The Bovine Tuberculosis Model for England and Wales (BoTMEW) focuses on the spread of Mycobacterium bovis, the causative agent of bovine tuberculosis, to individual herds by import of infected cattle, or by transmission from a local environmental reservoir, and on the detection of infected herds by surveillance which mainly relies on the single intradermal comparative cervical tuberculin skin test and detection at slaughter (Figure reproduced from Birch et al., 2018)
First I had to produce an alternative anonymised version of the model and its data that did not provide any personal information (e.g. reference to individual farms). Fortunately, the model already translated farm identities to arbitrary numbers for internal processing, so the model’s data could easily be fully anonymised by replacing all identifying information with fictional data, and removing all mentions of geographical locations with the exception of county. Checks demonstrated that the anonymised model’s outputs were a close match to the full model at county resolution. I reviewed the model with APHA’s Privacy Impact Assessment team to confirm that the anonymised model did not carry personal data.
The limit for supplementary material with a paper in BMC Veterinary Research is 20 MB, so my initial naïve plan was to collate a zip file of the model and its required data within that limit, to submit with the paper. Unfortunately, journals expect supplementary material to come as additional files, each of which is a single document. After an hour processing my zip file, the journal’s submission system listed about 1000 additional files that it had accepted from me. The submission system unpacked not just the zip file, but all the program libraries necessary to run the model. Clearly the journal’s provision for supplementary material could not provide the solution I needed.
Plan B was to publicly release the model, and add a reference to the repository where all the information and tools are held from within the paper. This would allow members of the public to freely use and distribute the model under the Open Government licence. A Data Object Identifier (DOI), which is a permanent unique electronic library ID (equivalent to the DOI used to permanently identify journal papers) is used to make the release indefinitely discoverable. The information labelled by a DOI cannot be changed, although it can be associated with explanatory information that updates over time. For example, the papers in a science journal all have DOI, which remain the same when the journal changes its name.
Another issue was the type of licence that could be used to enable the free distribution of an otherwise copyrighted work. Perhaps surprisingly, there are many variations of the conditions for public or open release of information, the most widespread and least restrictive of which is the Creative Commons 0 (CC0) license. However, work that falls under Crown copyright (which provides special copyright rules for materials created by civil servants, ministers and government departments and agencies) needs to be licensed through the Open Government Licence (currently version 3), which is maintained by the National Archives.
Zenodo
For most scientists the choice of data repository is self-evident they use specific repositories for the domains they work in, defined either by the field they work in, or the institution they work at. Veterinary epidemiologists working at government institutions are exceptions to this general rule. Within the Defra group, the Environmental Agency runs the interface and metadata catalogue to support data publication on data.gov.uk. However, that repository currently accepts only a limited type of data files which wouldn’t have catered for all my needs . There are relatively few general data repositories: ArXiv is used for documents and preprints, Dryad stores data for papers, has charges and only offers CC0 licences, Figshare is a commercial data repository that only offers CC0 licences, Dataverse is a framework for developing your own repository, the option I preferred was Zenodo.
BoTMEW as seen in the Zenodo open access directory which allows you to upload, describe and publish research outputs. We can see how many people have interacted with the BoTMEW files Colin has publically shared, including the number of downloads.
Zenodo is provided as a free open-access repository developed under the OpenAIRE program to support the European Research Council open access policies. It is operated by CERN, the European Organisation for Nuclear Research, using a small part of the spare space in their enormous high-performance computing infrastructure to promote the exchange of open data, software and other research related digital products. Zenodo has several useful features intended to facilitate the provision of open data within journal publications. Data and metadata can be saved in the repository, but only for private access until they are ready for general public release. DOI can be reserved before release, so that papers can be drafted with citations to data that will not be made available until the paper is published. The repository also provides versioning, which automatically links published papers to DOI for revisions of their supplementary data, even if the revisions are made after the papers are published.
So I am happy to report that the paper discussing the model outputs and their implications has been published open-access with the journal BMC Veterinary Research. The full model description is provided alongside as supplementary material. The model itself was released on Zenodo.

1 comment
Comment by 3win8 online game posted on
What's the difference between mass and burden? But what about places where cells don't get signal?
Interestingly enough, this all goes in order to personal
the liability.